
It will also discuss the applications of restriction enzymes in molecular biology and biotechnology and the potential limitations when working with these enzymes. This article will provide information about restriction enzymes, their properties, classification and action mechanisms. The discovery of restriction enzymes helped revolutionize the field of molecular biology by giving scientists the ability to manipulate DNA and study it in various applications including genetic engineering and DNA sequencing. Restriction enzymes are proteins produced by bacteria and archaea that can cleave DNA molecules at specific sites to create DNA fragments that have known sequences at each end. Type I, Type III and Type IV have more complex mechanisms of action and are less commonly used in research. It recognizes and cuts DNA at specific palindromic sequences, resulting in DNA fragments with sticky ends that can be easily ligated to other DNA fragments.
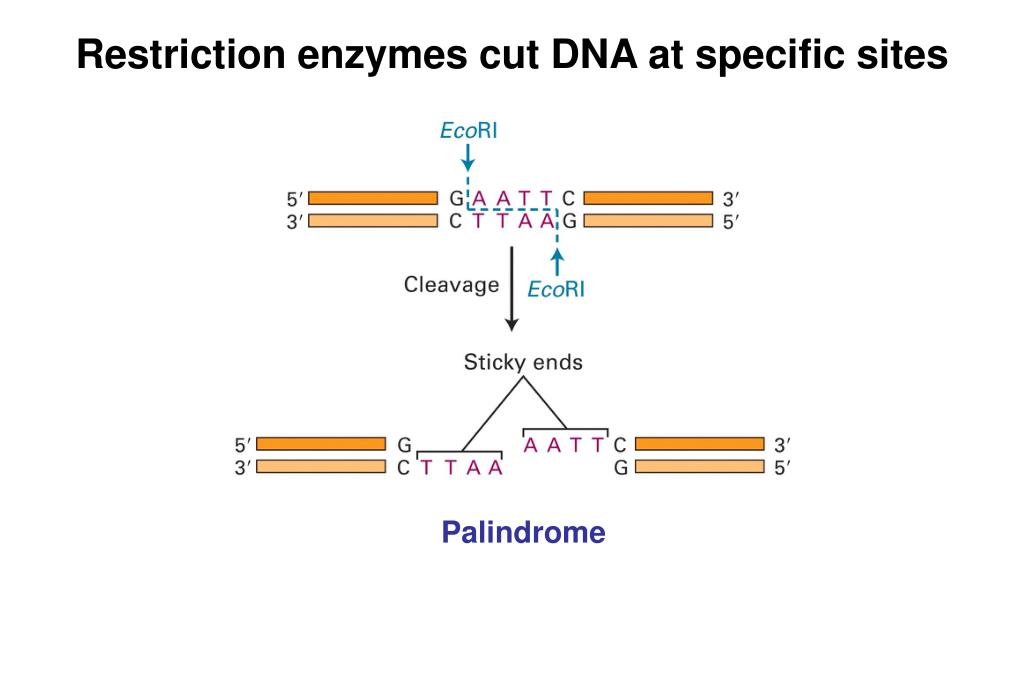
Type II is the most commonly used in molecular biology research. The four types of restriction enzymes are Type I, Type II, Type III and Type IV. HALO® Chromatography Columns and Consumables+. Sartorius laboratory instruments, consumables and services PerkinElmer - Innovating for a Healthier World Lab Thermometers & Temperature Measurement EquipmentĪgilent Chemistries and Supplies Portfolio
Quality Products from Sheldon Manufacturing Life Science Research Solutions, Products, and Resources VWR will support you from the latest life science products to the guaranteed purity of organic building blocks. Here, for example, the single difference in the sequences can be eliminated (red for blue or vice versa).A strong, vibrant research and development group is the lifeblood of all industries. Repairs can then be made (probably by the mechanism of homologous recombination). All that is needed is to form a loop so that the two sequences line up side-by-side. This orientation and redundancy may help ensure that a deleterious mutation in one copy of the set can be repaired using the information in another copy of that set. (The dashes represent the thousands of base pairs that separate adjacent palindromes.) The human Y chromosome contains 7 sets of genes - each set containing from 2 to 6 nearly-identical genes - oriented back-to-back or head-to-head that is, they are inverted repeats like the portion shown here.

Inverted repeats at either end of retroviral gene sequences aid in inserting the DNA copy into the DNA of the host.The DNA of many transposons is flanked by inverted repeats such as this one:ĥ' GGCCAGTCACAATGG.~400 nt.CCATTGTGACTGGCC 3'ģ' CCGGTCAGTGTTACC.~400 nt.GGTAACACTGACCGG 5'.Transcription factors are often dimers of identical proteins homodimers so it is not surprising that each member of the pair needs to "see" the same DNA sequence in the same orientation. The DNA sequence shown above is that of the glucocorticoid response element where n represents any nucleotide. The DNA to which transcription factors bind.In these cases, two different segments of the double helix read the same but in opposite directions.


 0 kommentar(er)
0 kommentar(er)
